Open Large Data
Fiji
Introduction
Datasets which are tens or hundreds of GB usually don’t fit into the RAM memory of a computer anymore. Therefore they require special steps when opening in Fiji.
We show three options to load and visualize the data in Fiji here:
- Load data virtually
- Load a subset of the data
- Visualize with BigDataViewer
Tip 1: Acquire also a small test data set when at the microscope (for example with 2 timepoints instead of 100). This test set will make it much less painful to develop an analysis workflow.
Tip 2: Use a workstation (like LIPS computers) which has larger RAM memory than your laptop. If you’re lucky the data now fits into RAM and you can directly open the image normally.
Step-by-step
Option 1: Load data virtually
Prerequisite: Bio-Formats Update site is activated (see general instructions).
Only the current slice is loaded into memory. This is good for previewing the full dataset quickly, but processing options are limited.
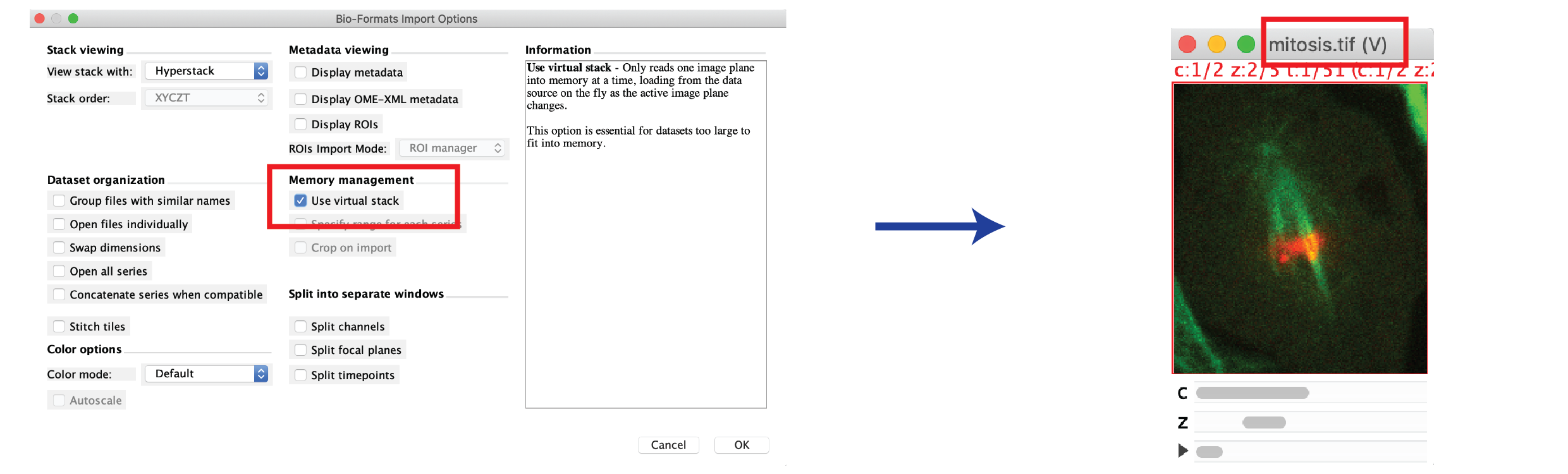
- Select File > Import > Bio-Formats, then select your image (or the master file, if a series).
- Check the “Use virtual stack” box, then press OK.
- The image title has a “(V)” appended which means that the image is loaded virtually.
- Examples of what can and cannot be done with virtual images:
- Max-Projection: Possible. The projected image must fit into memory.
- Filtering/Blurring: not possible.
- Copy a Roi region or substack: Possible, but the copied region is loaded into memory.
Option 2: Load a subset of the data
Prerequisite: Bio-Formats Update site is activated (see general instructions).
Data is cropped in space or time so that it fits into memory. This is very useful if a specimen is for example surrounded by lots of background. Or if the data can be processed sequentially, for example frame by frame.
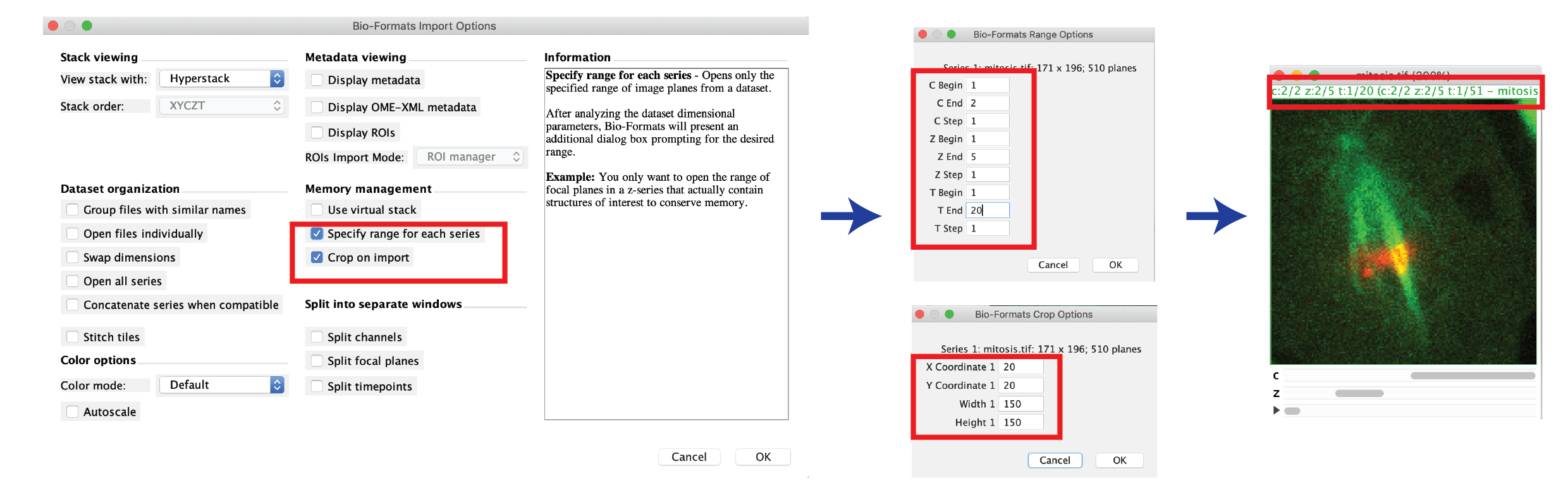
- Select File > Import > Bio-Formats, then select your image (or the master file, if a series).
- Check the “Specify range for each series” and/or “Crop on import” box, then press OK.
- Choose the channels, slices, frames and Roi region that you want to import, then press OK.
- The image is loaded into memory, and the reduced ranges can be seen in the image status bar.
Option 3: Visualize with BigDataViewer
Prerequisite: Bio-Formats Update site is activated (see general instructions).
The BigDataViewer (“BDV”) is very useful for exploring even very large datasets interactively and from any angle. Try it also especially when the virtual loading was too unresponsive. For actual image processing like thresholding you will however have to go back to “standard” ImageJ image windows.
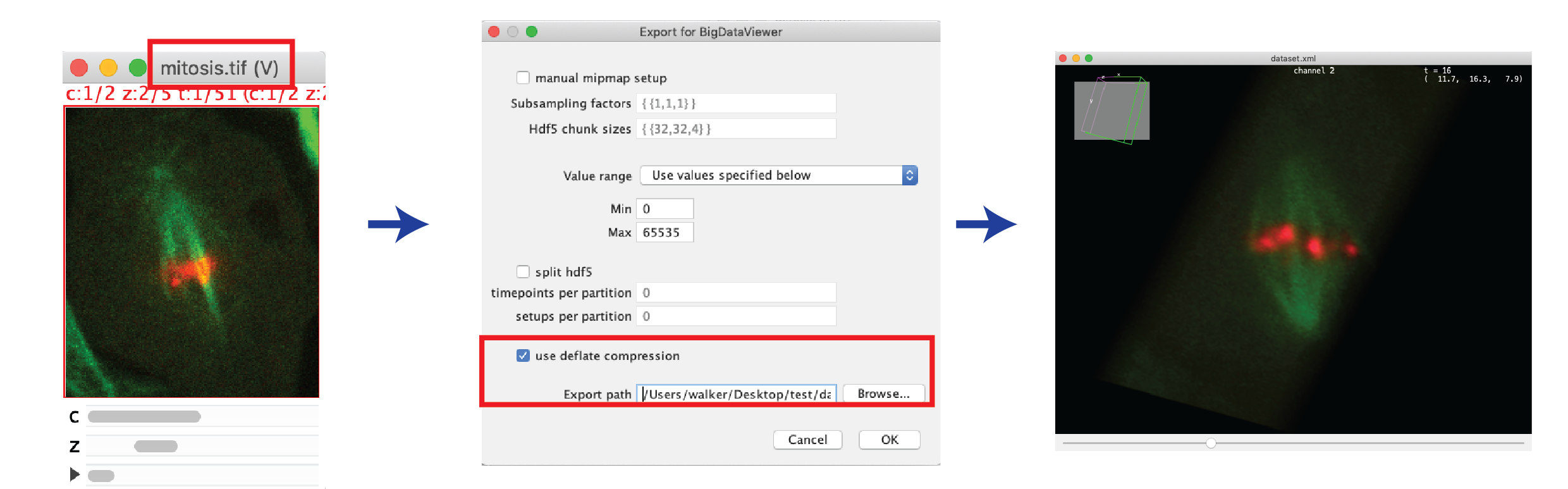
- For displaying in BDV, the dataset should be converted to HDF5 format (a hierarchical data format which allows for fast visualization) first:
- Open the image virtually with Bio-Formats, as described in Option 1.
- Make sure the image window is selected, then select: Plugins > BigDataViewer > Export Current Image as XML/HDF5.
- Define a save name, press OK, then wait until export is done.
- Open the resaved image with BigDataViewer: Plugins > BigDataViewer > Open XML/HDF5.
Quick usage instructions for BDV
BDV window must be selected for steps below:
- For help on navigation, bookmarking etc: Help > Show Help.
- To change channel brightness and color: Settings > Brightness & Color.
- Show all channels in multi-channel image: Settings > Visibility & Grouping, then check “enable fused mode”.
- Use bookmarking (see Help menu) to store positions and to go back to them. Also store the initial position to be able to reset the view if you lost the object.
- Save the settings: File > Save settings.
For detailed instructions check the BigDataViewer webpage.
Troubleshooting and more options
- If the export to XML/HDF5 causes trouble (is very slow), try exporting with the BigStitcher toolbox:
- Prerequisite: BigStitcher Update site is activated.
- Select Plugins > BigStitcher > Batch Processing > Define Dataset …
- Walk through the wizard, try to stick with defaults, resave as HDF5 (detailed instructions here).
- An exported XML/HDF5 dataset can also be opened via the BigStitcher plugin: Plugins > BigStitcher > BigStitcher.
- The viewing window is exactly the BigDataViewer.
- But there is an additional table for selecting timepoints, channels, etc..
Alternatives
Imaris
- Imaris can handle large data automatically. When opening your data in Imaris for the first time it gets automatically converted (copied) to the proprietary .ims format. This is a hierarchical data format which allows for responsive viewing of large data.